
Jung-Ja Kim, PhD
Emeritus Professor
Locations
- Biochemistry
TBRC C2960
Contact Information
Education
BS, Seoul National University
Biography
Dr. Kim received her PhD in Physical Chemistry from Cornell University, where she was a Fulbright Scholar. She conducted her postdoctoral study at Massachusetts Institute of Technology, where she was part of the research team under Dr. Alexander Rich that determined the first structure of transfer RNA. She joined the faculty of the Medical College of Wisconsin in 1978 and went on to establish the Structural Biology Research at MCW by setting up an X-ray Crystallographic Facility. Dr. Kim is a fellow of the American Association for the Advancement of Science (AAAS).
Research Interests
Our research interest is to study the structure-function relationship of biologically interesting molecules (enzymes, nucleic acids, receptors, etc.) using X-ray diffraction methods in combination with biochemical and computational methods. Our studies are focused on the following projects:
- Enzymes involved in fatty acid metabolism - Acyl-CoA dehydrogenases and related enzymes
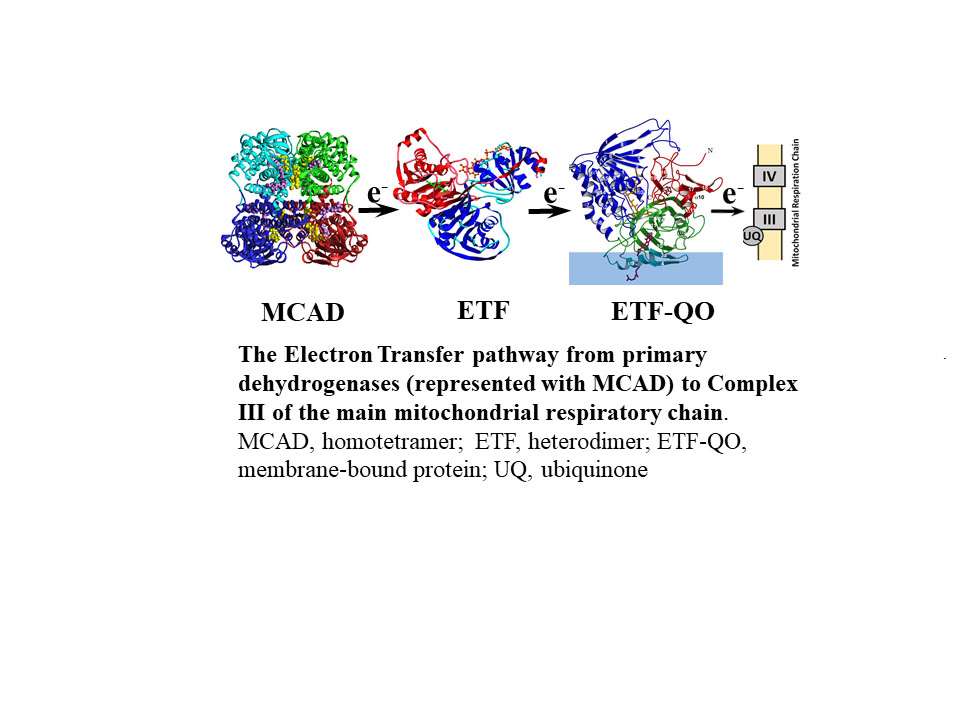
β-Oxidation of fatty acids is the principal energy-yielding process in the liver, heart, kidney, and muscle. It is carried out by a series of enzymes that successively and repetitively cleave acetyl-CoA fragments from fatty acyl-CoA molecules, until the fatty acyl-CoA is completely degraded. The rate of oxidation can be altered by diet (such as in starvation), physiological status (such as in pregnancy), and disease (such as in diabetes). The initial reaction in each cycle of fatty acid β-oxidation is the dehydrogenation of an acyl-CoA thioester to the corresponding trans-2,3-enoyl-CoA. This reaction is catalyzed by a family of closely related flavoenzymes, acyl-CoA dehydrogenases (ACADs), that exhibit distinct but overlapping chain length specificities for acyl-CoA thioesters.
Electron transfer from the dehydrogenases to the main mitochondrial respiratory chain is catalyzed, in sequence, by electron transfer flavoprotein (ETF) and the membrane-bound ETF-ubiquinone oxidoreductase (ETF-QO), an iron-sulfur flavoprotein. We have determined the structures of several acyl-CoA dehydrogenases, including medium chain acyl-CoA dehydrogenase (MCAD, the most abundant ACAD), human ETF, and ETF-QO. These structures have enabled us to study the molecular basis of electron transfer between the dehydrogenases and ETF and between ETF and ETF-QO, and flavoprotein-flavoprotein interactions in general.
MCAD deficiency is the most common among all fatty acid metabolism disorders. It is manifested by fasting intolerance, hypoglycemic coma, failure of ketogenesis, and dicarboxylic aciduria, and has been implicated in sudden infant death syndrome.
- NADPH-cytochrome P450 reductase (CYPOR)
NADPH-cytochrome P450 oxidoreductase (CYPOR) is a microsomal diflavin protein that shuttles electrons from NADPH → FAD → FMN to members of the ubiquitous cytochrome P450 superfamily. In humans, the cytochromes P450 (cyt P450) are one of the most important families of proteins involved in the biosynthesis and degradation of a vast number of endogenous compounds and the detoxification and biodegradation of most foreign compounds. We have determined the structure of human CYPOR. CYPOR undergoes a large conformational change in the course of electron transfer from FAD to FMN and from FMN to cyt P450, its physiological redox partner. Furthermore, as CYPOR is the prototype of the diflavin enzyme family that utilizes pyridine nucleotides, our findings can be applied to the mechanism of electron transfer in other members of the family, including NOS isozymes and methionine synthase reductase.
NADPH-cytochrome P450 reductase dancing with cytochrome P450 Movie
- Mannose 6-Phosphate Receptors (MPRs)
MPRs are responsible for the targeting of lysosomal acid hydrolases to lysosomes. In collaboration with Dr. Nancy Dahms, we have been studying the structure/function relationships of these receptors. We have solved the structures of the cation-dependent MPR (CD-MPR) with and without mannose 6-phosphate ligand and complexes with high mannose oligosaccharides. We are extending our studies to the larger of the two receptors, the insulin-like growth factor II/cation-independent MPR (IGF-II/CI-MPR).
Publications
-
(Narayanan B, Xia C, McAndrew R, Shen AL, Kim JP.) Sci Rep. 2024 Jun 05;14(1):12976 PMID: 38839792 PMCID: PMC11153573 06/06/2024
-
(Narayanan B, Xia C, McAndrew R, Shen AL, Kim JP.) Res Sq. 2024 Feb 29 PMID: 38464032 PMCID: PMC10925408 03/11/2024
-
(Xia C, Lou B, Fu Z, Mohsen AW, Shen AL, Vockley J, Kim JP.) iScience. 2021 Oct 22;24(10):103153 PMID: 34646991 PMCID: PMC8497999 10/15/2021
-
(Hubbard PA, Xia C, Shen AL, Kim JP.) Arch Biochem Biophys. 2021 Apr 15;701:108792 PMID: 33556357 PMCID: PMC8020834 02/09/2021
-
(Olson LJ, Misra SK, Ishihara M, Battaile KP, Grant OC, Sood A, Woods RJ, Kim JP, Tiemeyer M, Ren G, Sharp JS, Dahms NM.) Commun Biol. 2020 Sep 09;3(1):498 PMID: 32908216 PMCID: PMC7481795 SCOPUS ID: 2-s2.0-85090384984 09/11/2020
-
(Xia C, Shen AL, Duangkaew P, Kotewong R, Rongnoparut P, Feix J, Kim JP.) Biochemistry. 2019 May 14;58(19):2408-2418 PMID: 31009206 PMCID: PMC6873807 SCOPUS ID: 2-s2.0-85065650755 04/23/2019
-
Crystal structure of human mitochondrial trifunctional protein, a fatty acid β-oxidation metabolon.
(Xia C, Fu Z, Battaile KP, Kim JP.) Proc Natl Acad Sci U S A. 2019 Mar 26;116(13):6069-6074 PMID: 30850536 PMCID: PMC6442613 03/10/2019
-
(Xia C, Rwere F, Im S, Shen AL, Waskell L, Kim JP.) Biochemistry. 2018 Feb 13;57(6):945-962 PMID: 29308883 PMCID: PMC5967631 01/09/2018
-
(Galiakhmetov AR, Kovrigina EA, Xia C, Kim JP, Kovrigin EL.) J Biomol NMR. 2018 Jan;70(1):21-31 PMID: 29168021 PMCID: PMC5820150 11/24/2017
-
(Kovrigina EA, Pattengale B, Xia C, Galiakhmetov AR, Huang J, Kim JP, Kovrigin EL.) Biochemistry. 2016 Nov 01;55(43):5973-5976 PMID: 27741572 PMCID: PMC6505697 11/02/2016
-
(McCammon KM, Panda SP, Xia C, Kim JJ, Moutinho D, Kranendonk M, Auchus RJ, Lafer EM, Ghosh D, Martasek P, Kar R, Masters BS, Roman LJ.) J Biol Chem. 2016 Sep 23;291(39):20487-502 PMID: 27496950 PMCID: PMC5034044 08/09/2016
-
Mitochondrial Protein Interaction Mapping Identifies Regulators of Respiratory Chain Function.
(Floyd BJ, Wilkerson EM, Veling MT, Minogue CE, Xia C, Beebe ET, Wrobel RL, Cho H, Kremer LS, Alston CL, Gromek KA, Dolan BK, Ulbrich A, Stefely JA, Bohl SL, Werner KM, Jochem A, Westphall MS, Rensvold JW, Taylor RW, Prokisch H, Kim JP, Coon JJ, Pagliarini DJ.) Mol Cell. 2016 Aug 18;63(4):621-632 PMID: 27499296 PMCID: PMC4992456 08/09/2016

