Emma Morrison, PhD
Associate Professor
Locations
- TBRC C2980
Contact Information
General Interests
Education
PhD, Washington University in St. Louis, 2014
BA, Johns Hopkins University, 2008
Biography
Research Interests
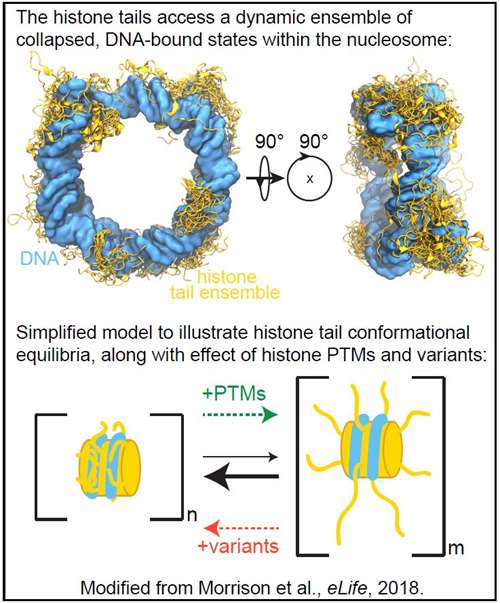
The Morrison lab is interested in understanding molecular mechanisms of chromatin regulation, which are key in fundamental mechanisms of gene regulation and provide valuable insight into human health and disease. A complex network of machinery dynamically regulates the organization and accessibility of the human genome within chromatin. This re-organization starts at the level of the nucleosome, the basic subunit of chromatin. The nucleosome is a histone protein-DNA complex, and the N-terminal tails of the histone proteins protrude from the core complex to interact with regulatory machinery. These interactions are often driven by specific histone post-translational modifications (PTMs).
Our recent studies have contributed to recognizing that the histone tails have reduced accessibility to interactions with chromatin regulatory machinery within the nucleosome due to interactions with DNA. These findings suggest that there are other nuclear factors in vivo that modulate the conformation and in turn the accessibility of the tails to regulate interactions with protein machinery that lead to downstream chromatin regulation. We are taking a quantitative, biophysical approach in order to determine the role of the histone tails in chromatin structure and dynamics by studying the conformation and dynamics of the histone tails within nucleosomes. Factors such as histone PTMs and histone variants have the potential to directly regulate histone tail conformation, thereby regulating chromatin structure and modulating accessibility to binding factors.
The lab uses NMR spectroscopy along with a range of other biophysical and biochemical techniques in order to investigate these questions. If you are interested in joining the team, please contact Dr. Morrison.
Publications
-
Multidrug Resistance Transporter EmrE is Only Moderately Sensitive to Lipid Composition.
(Hiett AB, Hisao GS, Robinson AE, Morrison EA, Henzler-Wildman KA.) Biophys J. 2026 Jan 28 PMID: 41612699 01/30/2026
-
(Li Y, Xie N, Chen R, Lee AR, Lovnicki J, Morrison EA, Fazli L, Zhang Q, Musselman CA, Wang Y, Huang J, Gleave ME, Collins C, Dong X.) Eur Urol. 2025 Jul;88(1):121 PMID: 40340107 SCOPUS ID: 2-s2.0-105004587457 05/09/2025
-
Histone H3 tail charge patterns govern nucleosome condensate formation and dynamics.
(Hammonds EF, Singh A, Suresh KK, Yang S, Zahorodny SSM, Gupta R, Potoyan DA, Banerjee PR, Morrison EA.) bioRxiv. 2025 Apr 10 PMID: 40291647 PMCID: PMC12027143 04/28/2025
-
Arginine anchor points govern H3 tail dynamics.
(Jennings CE, Zoss CJ, Morrison EA.) Front Mol Biosci. 2023;10:1150400 PMID: 37261328 PMCID: PMC10228543 06/01/2023
-
Nucleosome Core Particle Reconstitution with Recombinant Histones and Widom 601 DNA.
(Hammonds EF, Morrison EA.) Methods Mol Biol. 2023;2599:177-190 PMID: 36427150 SCOPUS ID: 2-s2.0-85142940540 11/26/2022
-
(Paintsil EA, Morrison EA.) Methods Mol Biol. 2023;2599:163-175 PMID: 36427149 SCOPUS ID: 2-s2.0-85142940771 11/26/2022
-
Histone H3 and H4 tails play an important role in nucleosome phase separation.
(Hammonds EF, Harwig MC, Paintsil EA, Tillison EA, Hill RB, Morrison EA.) Biophys Chem. 2022 Apr;283:106767 PMID: 35158124 PMCID: PMC8963862 SCOPUS ID: 2-s2.0-85124393236 02/15/2022
-
(Lupo BE, Chu P, Harms MJ, Morrison EA, Musselman CA.) J Mol Biol. 2021 Jul 09;433(14):166845 PMID: 33539881 PMCID: PMC8184587 SCOPUS ID: 2-s2.0-85101513034 02/05/2021
-
Nucleosome composition regulates the histone H3 tail conformational ensemble and accessibility.
(Morrison EA, Baweja L, Poirier MG, Wereszczynski J, Musselman CA.) Nucleic Acids Res. 2021 May 07;49(8):4750-4767 PMID: 33856458 PMCID: PMC8096233 SCOPUS ID: 2-s2.0-85106069963 04/16/2021
-
RNA Splicing of the BHC80 Gene Contributes to Neuroendocrine Prostate Cancer Progression.
(Li Y, Xie N, Chen R, Lee AR, Lovnicki J, Morrison EA, Fazli L, Zhang Q, Musselman CA, Wang Y, Huang J, Gleave ME, Collins C, Dong X.) Eur Urol. 2019 Aug;76(2):157-166 PMID: 30910347 SCOPUS ID: 2-s2.0-85063207106 03/27/2019
-
The C terminus of the bacterial multidrug transporter EmrE couples drug binding to proton release.
(Thomas NE, Wu C, Morrison EA, Robinson AE, Werner JP, Henzler-Wildman KA.) J Biol Chem. 2018 Dec 07;293(49):19137-19147 PMID: 30287687 PMCID: PMC6295725 SCOPUS ID: 2-s2.0-85058168173 10/06/2018
-
Reading More than Histones: The Prevalence of Nucleic Acid Binding among Reader Domains.
(Weaver TM, Morrison EA, Musselman CA.) Molecules. 2018 Oct 12;23(10) PMID: 30322003 PMCID: PMC6222470 SCOPUS ID: 2-s2.0-85054890627 10/17/2018