
Jimmy B. Feix, PhD
Professor
Locations
- Biophysics
- MFRC 2040B
Contact Information
Education
PhD, Chemistry, University of Kentucky, Lexington, KY, 1981
BS, Chemistry, Western Kentucky University, Bowling Green, KY, 1976
Biography
I received my BS in chemistry from Western Kentucky University and PhD in chemistry from University of Kentucky. I completed my postdoctoral training in membrane biophysics at MCW. I was appointed as MCW faculty in 1985.
Research Interests
My current research interests are in the use of advanced electron paramagnetic resonance (EPR) spin labeling techniques to study the mechanisms that drive peptide – membrane and protein – membrane interactions, and in the development of new peptide antibiotics.
We utilize site-directed spin labeling (SDSL) EPR spectroscopy and other biophysical and biochemical techniques to work on problems related to the membrane interactions of peptides and proteins and the structure and dynamics of membrane proteins.
Mechanism of Activation and Membrane Interactions of Pseudomonas toxin ExoU
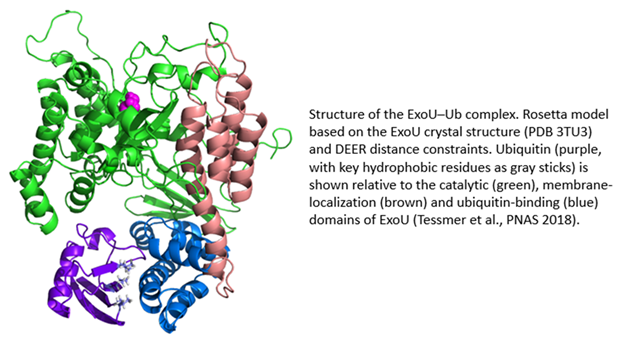
ExoU is a 74 kDa protein produced by the Gram-negative bacterial pathogen, Pseudomonas aeruginosa. ExoU is injected directly into eukaryotic cells by a needle-like Type III secretion system (T3SS). Once inside the eukaryotic cell, ExoU functions as a phospholipase to disrupt membranes and facilitate dissemination of the bacterial infection. ExoU also serves as a model system for the study of protein–membrane and protein–protein interactions. In a novel regulatory mechanism, ExoU is activated through protein–protein interaction with ubiquitin (Ub). Since Ub is exclusively a eukaryotic protein, this prevents the ExoU toxin from being activated within the bacterium. Using a combination of site-directed spin labeling EPR and double electron-electron resonance (DEER) spectroscopy, supported by mutagenesis and biochemical studies, we have identified the ExoU–Ub binding interface. Additional EPR studies in conjunction with computational analysis have led to a model for the membrane-bound ExoU-Ub complex. The goals of these studies are to facilitate the development of novel inhibitors of ExoU as a means to attenuate the virulence of P. aeruginosa infections.

Mechanisms of Antimicrobial Peptides: Membrane Interactions
Antibiotic resistance is an increasingly serious problem in the treatment of infectious disease. During the past two decades, a large number of peptides with potent antibacterial, antiviral, and antifungal properties have been identified from a wide range of both vertebrate and invertebrate species. These antimicrobial peptides (AMPs) are an important component of the innate arm of host resistance, serving as a first line of defense against infection. Despite being evolutionarily ancient, resistance to AMPs has only rarely been observed. Consequently, there is great interest in the development of these peptides for the treatment of drug-resistant infections.

Models of Transmembrane Channel Formation

Our laboratory is involved in elucidating the mechanisms by which AMPs disrupt bacterial membrane structure, determining the basis of AMP selectivity for microbial cells, and developing more effective antimicrobial peptides and peptidomimetics. Whereas classical antibiotics generally target cell wall synthesis, protein translation, or some other highly specific target, AMPs are believed to function by directly disrupting the microbial cell membrane. Peptides are prepared using either recombinant DNA methods or solid-phase chemical synthesis, and their interactions with model membranes (liposomes) and cells are characterized by a variety of physical techniques including circular dichroism, fluorescence, and EPR site-directed spin labeling (SDSL). Our fundamental hypothesis is that a more complete understanding of peptide structure and dynamic interactions with the membrane will allow the design and development of improved AMPs and related antibiotics for the treatment of infections by existing drug-resistant strains such as Pseudomonas aeruginosa and methicillin-resistant Staphylococcus aureus (MRSA).
Membrane Proteins I (PDF)
Membrane Proteins II (PDF)
Publications
-
Microbial Metabolism of Levodopa as an Adjunct Therapeutic Target in Parkinson's Disease.
(Feix JB, Cheng G, Hardy M, Kalyanaraman B.) Antioxidants (Basel). 2026 Jan 17;15(1) PMID: 41596178 PMCID: PMC12838373 01/28/2026
-
(Cheng G, Hardy M, Feix JB, Kalyanaraman B.) Sci Rep. 2025 Jun 06;15(1):20004 PMID: 40481059 PMCID: PMC12144245 SCOPUS ID: 2-s2.0-105007531272 06/07/2025
-
(Cheng G, Hardy M, Hillard CJ, Feix JB, Kalyanaraman B.) Commun Biol. 2024 May 30;7(1):668 PMID: 38816577 PMCID: PMC11139878 SCOPUS ID: 2-s2.0-85194991828 05/31/2024
-
(Gies SL, Tessmer MH, Frank DW, Feix JB.) Appl Magn Reson. 2024 Mar;55(1-3):279-295 PMID: 39175603 PMCID: PMC11340903 08/23/2024
-
(Gies SL, Tessmer MH, Frank DW, Feix JB.) Applied Magnetic Resonance. March 2024;55(1-3):279-295 SCOPUS ID: 2-s2.0-85173909117 03/01/2024
-
Characterization of the ExoU activation mechanism using EPR and integrative modeling.
(Tessmer MH, DeCero SA, Del Alamo D, Riegert MO, Meiler J, Frank DW, Feix JB.) Sci Rep. 2020 Nov 12;10(1):19700 PMID: 33184362 PMCID: PMC7665212 SCOPUS ID: 2-s2.0-85095885803 11/14/2020
-
Rapid Simulation of Unprocessed DEER Decay Data for Protein Fold Prediction.
(Del Alamo D, Tessmer MH, Stein RA, Feix JB, Mchaourab HS, Meiler J.) Biophys J. 2020 Jan 21;118(2):366-375 PMID: 31892409 PMCID: PMC6976798 SCOPUS ID: 2-s2.0-85077146138 01/02/2020
-
(Springer TI, Reid TE, Gies SL, Feix JB.) J Biol Chem. 2019 Dec 13;294(50):19012-19021 PMID: 31662432 PMCID: PMC6916483 SCOPUS ID: 2-s2.0-85076501851 10/31/2019
-
(Xia C, Shen AL, Duangkaew P, Kotewong R, Rongnoparut P, Feix J, Kim JP.) Biochemistry. 2019 May 14;58(19):2408-2418 PMID: 31009206 PMCID: PMC6873807 SCOPUS ID: 2-s2.0-85065650755 04/23/2019
-
(Feix JB, Kohn S, Tessmer MH, Anderson DM, Frank DW.) Cell Biochem Biophys. 2019 Mar;77(1):79-87 PMID: 30047043 PMCID: PMC6347562 SCOPUS ID: 2-s2.0-85050694302 07/27/2018
-
Identification of a ubiquitin-binding interface using Rosetta and DEER.
(Tessmer MH, Anderson DM, Pickrum AM, Riegert MO, Moretti R, Meiler J, Feix JB, Frank DW.) Proc Natl Acad Sci U S A. 2018 Jan 16;115(3):525-530 PMID: 29295930 PMCID: PMC5776994 SCOPUS ID: 2-s2.0-85042128224 01/04/2018
-
(Herneisen AL, Sahu ID, McCarrick RM, Feix JB, Lorigan GA, Howard KP.) Biochemistry. 2017 Nov 07;56(44):5955-5963 PMID: 29034683 PMCID: PMC6112238 SCOPUS ID: 2-s2.0-85033361759 10/17/2017

