
Michaela Patterson, PhD
Associate Professor
Locations
- Cell Biology, Neurobiology and Anatomy
MEB M4575
Cardiovascular Research Center
Contact Information
Education
PhD, University of California, Los Angeles, CA
BS, Bates College, Lewiston, ME
Research Interests
Our lab’s scientific interests lie at the intersection of cellular ploidy, genetic diversity, basal heart physiology, and tissue repair. We employ a mix of forward genetics approaches and resources to identify novel players in the traits we are exploring, along with classic reverse genetics and engineered animal models to assess causative relationships between identified genes and correlated phenotypes. Some specific areas of interest include:
1) Validation of the genetic underpinnings of cardiomyocyte ploidy and heart regeneration.
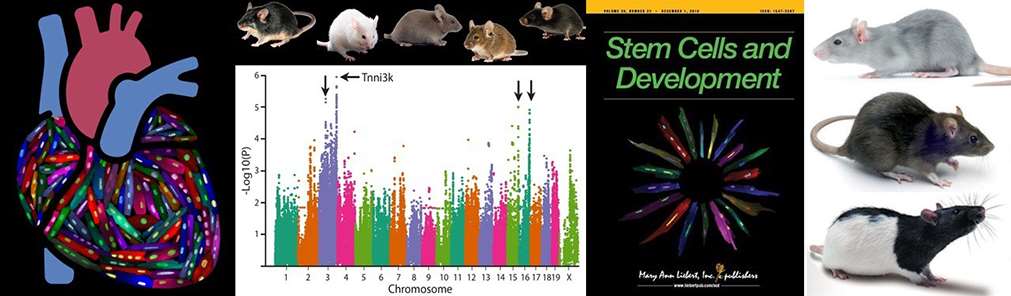
Patterson et al., Nature Genetics 2017 examined the relationship between cardiomyocyte ploidy and myocardial regeneration. Using a collection of 120 inbred mouse strains known as the Hybrid Mouse Diversity Panel, we uncovered multiple genetic loci associated with cardiomyocyte ploidy. We went on to validate the causative gene from the first locus, which proved to be Tnni3k.
The Patterson Lab has multiple projects underway (along with others still looking for a project lead) surrounding the other loci to arise from this screen. Using a variety of reverse genetic tools, including genetically engineered mice, viral induction, and more, some of the questions we commonly ask are:
- Is a candidate gene in the locus responsible for cardiomyocyte ploidy phenotypes AND what role do they play post-MI?
- Do the candidate genes have “functional” variants and what effect do they have on the gene’s molecular function and on ploidy?
- How do multiple genes, which individually influence ploidy, act together?
- Not all candidate genes function in cardiomyocytes, but instead act indirectly through another cell. What other cell types influence cardiomyocyte ploidy and MI outcomes?
2) New genetic screens to identify more players in cardiomyocyte polyploidization and outcomes after heart attack.
Our past work utilized the Hybrid Mouse Diversity Panel, a collection of inbred mouse strains designed to support genome-wide association studies. Our colleagues here at the Medical College of Wisconsin have recently rederived the rat equivalent resource - the Hybrid Rat Diversity Panel. The rat panel has many benefits that make it a superior genetic resource to the mouse panel, most notably that it is four times more genetically diverse. We have multiple projects underway, many of them collaborative, to examine a multitude of phenotypes across the Rat Panel. Phenotypes range from assessment of relevant cell populations to basal heart physiology to outcomes after heart attack and more. This arm of the Patterson lab has endless possibilities for expansion.
3) Polyploidy in other cell types.
Cardiomyocytes are not the only somatic cell in the body that displays substantial polyploidy. By examining the frequency of polyploid cells in other tissues we hope to better understand the overarching function of polyploidy in cell biology and determine if cell types utilize overlapping or distinct molecular mechanisms to drive polyploidization.
4) Bring your own ideas!
We are actively recruiting trainees at the graduate student and postdoctoral levels. We welcome new ideas to transform the way we think about tissue regeneration an repair. If you are interested in joining or rotating in the Patterson Lab, please contact Michaela Patterson, PhD, at mpatterson@mcw.edu.
Publications
-
Cardiomyocyte ploidy is dynamic during postnatal development and varies across genetic backgrounds.
(Swift SK, Purdy AL, Kolell ME, Andresen KG, Lahue C, Buddell T, Akins KA, Rau CD, O'Meara CC, Patterson M.) Development. 2023 Apr 01;150(7) PMID: 36912240 PMCID: PMC10113957 SCOPUS ID: 2-s2.0-85152488769 03/14/2023
-
Measuring cardiomyocyte cell-cycle activity and proliferation in the age of heart regeneration.
(Auchampach J, Han L, Huang GN, Kühn B, Lough JW, O'Meara CC, Payumo AY, Rosenthal NA, Sucov HM, Yutzey KE, Patterson M.) Am J Physiol Heart Circ Physiol. 2022 Apr 01;322(4):H579-H596 PMID: 35179974 PMCID: PMC8934681 SCOPUS ID: 2-s2.0-85127729067 02/19/2022
-
(Wang X, Wan TC, Lauth A, Purdy AL, Kulik KR, Patterson M, Lough JW, Auchampach JA.) J Mol Cell Cardiol. 2022 Feb;163:9-19 PMID: 34610340 PMCID: PMC8816866 SCOPUS ID: 2-s2.0-85116932867 10/06/2021
-
IL4Rα signaling promotes neonatal cardiac regeneration and cardiomyocyte cell cycle activity.
(Paddock SJ, Swift SK, Alencar-Almeida V, Kenarsary A, Alvarez-Argote S, Flinn MA, Patterson M, O'Meara CC.) J Mol Cell Cardiol. 2021 Dec;161:62-74 PMID: 34343540 PMCID: PMC8629844 SCOPUS ID: 2-s2.0-85112431618 08/04/2021
-
(Wang X, Lupton C, Lauth A, Wan TC, Foster P, Patterson M, Auchampach JA, Lough JW.) J Mol Cell Cardiol. 2021 Jun;155:88-98 PMID: 33609538 PMCID: PMC8154663 SCOPUS ID: 2-s2.0-85102458794 02/21/2021
-
Introductions to the Community: Early-Career Researchers in the Time of COVID-19.
(.) Cell Stem Cell. 2020 Jul 02;27(1):13-14 PMID: 32619510 PMCID: PMC7331526 07/04/2020
-
(Gan P, Patterson M, Watanabe H, Wang K, Edmonds RA, Reinholdt LG, Sucov HM.) Sci Rep. 2020 May 05;10(1):7605 PMID: 32371981 PMCID: PMC7200697 05/07/2020
-
(Shen H, Gan P, Wang K, Darehzereshki A, Wang K, Kumar SR, Lien CL, Patterson M, Tao G, Sucov HM.) Elife. 2020 Mar 13;9 PMID: 32167474 PMCID: PMC7105374 03/14/2020
-
Cardiomyocyte Polyploidy and Implications for Heart Regeneration.
(Gan P, Patterson M, Sucov HM.) Annu Rev Physiol. 2020 Feb 10;82:45-61 PMID: 31585517 10/06/2019
-
(Patterson M, Swift SK.) Stem Cells Dev. 2019 Dec 01;28(23):1527-1539 PMID: 31608782 PMCID: PMC11001963 10/15/2019
-
Tnni3k alleles influence ventricular mononuclear diploid cardiomyocyte frequency.
(Gan P, Patterson M, Velasquez A, Wang K, Tian D, Windle JJ, Tao G, Judge DP, Makita T, Park TJ, Sucov HM.) PLoS Genet. 2019 Oct;15(10):e1008354 PMID: 31589606 PMCID: PMC6797218 10/08/2019
-
Frequency of mononuclear diploid cardiomyocytes underlies natural variation in heart regeneration.
(Patterson M, Barske L, Van Handel B, Rau CD, Gan P, Sharma A, Parikh S, Denholtz M, Huang Y, Yamaguchi Y, Shen H, Allayee H, Crump JG, Force TI, Lien CL, Makita T, Lusis AJ, Kumar SR, Sucov HM.) Nat Genet. 2017 Sep;49(9):1346-1353 PMID: 28783163 PMCID: PMC5736145 08/08/2017

